A new twist in protein engineering points toward smarter medicines that know where to act and when. University of Washington researchers report a protein toolkit that uses Boolean logic to steer therapies to the right place, improving targeted delivery while reducing off target effects. The study appears in Nature Chemical Biology, with experiments spanning purified proteins, hydrogels, and living mammalian cells.
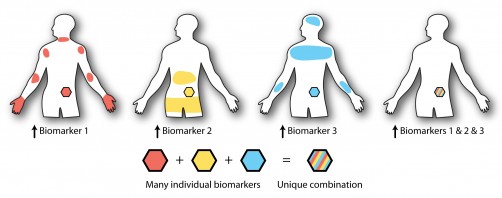
The core idea is simple to picture. Imagine a city map dotted with landmarks, each landmark a biomarker such as a protease. Only certain neighborhoods contain the right combination. The team built protein cargos with tail pieces that fold into specific topologies and cleave only when the correct inputs are present. In practice, that means a therapy can check for one signal, or two in parallel, or three in series, and act only if the Boolean rule evaluates to yes.
From Chemistry Bottlenecks To Genetic Assembly
Earlier logic-responsive materials demanded painstaking, low yield organic synthesis. The UW group replaced that bottleneck with recombinant expression and tag catcher ligations, letting living cells assemble the topology automatically. They compiled an entire library of 17 operators that implement the full set of YES, OR, and AND combinations for three orthogonal protease inputs, then validated each operator’s truth table in solution, on magnetic beads, and inside soft hydrogel networks.
The shift is not just elegant, it is scalable. Proteins were produced in weeks, not months, and with yields suitable for iterative design. As senior author Cole DeForest put it in the university release:
“We’ve now finally figured out how to produce these systems faster, at scale and with dramatically enhanced logical complexity. We are excited about how these will lead to more sophisticated and scalable disease-honing therapies.”
The team further demonstrated multiplexing. In one hydrogel, three different fluorescent cargos were tethered by different logic gates, then released independently when their specific input combinations were applied. They also built a five-input operator that responded only when a composite condition across five distinct proteases was met, a level of complexity unusual for material platforms and achieved here through genetic encodability.
Cells As Both Factory And Testbed
Logic did not stop at test tubes. The researchers attached a logic-gated nanobody to label HER2 on breast cancer cells only under preprogrammed conditions. They then moved the computation inside cells. A stable HEK293 line expressed a membrane-anchored fluorescent protein bridged by an OR over AND gate. When cells produced the matching proteases, the bridge was cut and the cargo moved from membrane to cytosol, exactly as the truth table predicted.
The scene in the microscope is clear and visual. A thin green rim hugs the cell’s edge at baseline. Supply the correct inputs, and the rim dissolves into an even glow across the cytoplasm, a spatial readout of a logical decision. That spatial control hints at therapies that arm or disarm only within target tissues, or even within subcellular compartments.
The enabling parts list will be familiar to protein engineers: split intein cyclization to create closed loops, orthogonal Spy, Snoop, and Dog Tag Catcher ligations to form branched or bicyclic topologies, and a set of proteases chosen to avoid crosstalk. The power comes from nesting these pieces into minimal gate motifs that can be layered without losing signal to noise.
Cost and speed matter for translation. Recombinant expression in bacteria, followed by standard purification and size exclusion, delivered clean constructs at useful scale. That design build test loop, now measured in days to weeks, makes it realistic to tune linkers, swap inputs, and debug leakage. As co first author Murial Ross said:
“The sky’s the limit. You can create delayed and independent delivery of many different components in one treatment.”
Applications are obvious and near term. Oncology tops the list, where combinations of proteases and microenvironmental cues can serve as addresses. Diagnostics are plausible too, such as blood tests that turn on only when a complex molecular pattern is present. Because the logic is encoded in amino acid sequences, it should be straightforward to retarget gates toward disease relevant inputs or to couple them to non protease sensors that map light, small molecules, or redox to protease activity.
There are caveats. Higher order constructs can be harder to purify to absolute homogeneity, and in cell expression can create timing issues if proteases clip linkers before cyclization completes. Still, the group’s data show crisp truth tables across many contexts, suggesting that careful topology design can keep noise low.
If these proteins can chaperone real payloads with the same fidelity seen in fluorescent demos, clinicians may gain a new set of switches to control where a drug goes and when it acts, reducing dose while increasing effect. Logic at the molecular scale is no longer a metaphor. It is a blueprint a cell can build.
Nature Chemical Biology: 10.1038/s41589-025-02037-5
If our reporting has informed or inspired you, please consider making a donation. Every contribution, no matter the size, empowers us to continue delivering accurate, engaging, and trustworthy science and medical news. Independent journalism requires time, effort, and resources—your support ensures we can keep uncovering the stories that matter most to you.
Join us in making knowledge accessible and impactful. Thank you for standing with us!